Attachment A
ATTACHMENT A. A Report of Expert Consultations on Rapid Molecular Testing to Detect Drug Resistant Tuberculosis in the United States.docx
Surveys of State, Tribal, Local and Territorial (STLT) Governmental Health Agencies
ATTACHMENT A
OMB: 0920-0879
ATTACHMENT A:

![]()
This report is based on contributions of an expert panel of consultants (E Desmond PhD, California Dept. of Public Health; K Field RN MSN, Washington Dept. of Health; P Griffin BBA, Kansas Dept. of Health and Environment; D Ingman BS MT DPHHS, Montana Dept. of Public Health and Human Services; K Musser PhD, New York State Dept. of Health; M Oxtoby MD, New York State Dept. of Health; V Pruitt, Alabama Dept. of Public Health; J Razeq PhD, Maryland Dept. of Health; M Salfinger MD, Florida Dept. of Health; B Seaworth MD, Heartland National TB Training and Education Center; J Watt MD MPH, California Dept. of Public Health; K Wroblewski MPH, Association of Public Health Laboratories; S Zanto CLS(NCA) SM(NRM), Montana Public Health Laboratory) and CDC participants (K Ijaz MD, B Metchock DrPH, J Posey PhD, A Starks PhD, and T Shinnick PhD, Division of Tuberculosis Elimination, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention).
Contents Page
Executive summary 1
Introduction 2
Background on tests for molecular detection of drug resistance (DR) 3
General considerations and principles for a molecular DR testing service 4
Possible scenarios and scope of testing for a molecular DR testing service 8
Research needs 9
General recommendations of the Expert Panel 9
Communication plan for the report 12
Recommendations 12
References 13
Panel members and CDC participants 14
Appendices
Molecular basis of drug resistance and molecular DR tests 16
Roles and responsibilities in a regional laboratory system for 18
providing for molecular DR testing for TB
Flow chart of steps in a molecular DR testing service 23
Executive summary
Rapid drug-susceptibility tests are a pressing public health and diagnostic need because of the rise in multidrug-resistant and extensively drug-resistant tuberculosis (MDR/XDR TB) globally (1). Published studies (2,3) suggest that, compared to conventional culture-based methods, the rapid detection of rifampin-resistance using molecular methods can enable earlier initiation of effective therapy and thereby reduce periods of infectiousness of MDR TB cases by as much as six weeks and improve patient outcomes; both of which may have a large impact on efforts to control MDR TB. It is estimated that preventing a single case of MDR TB would save the U.S. health care system more than $250,000 (4). In recognition of the importance of rapid drug-susceptibility testing, the Advisory Council for the Elimination of Tuberculosis (ACET) passed a resolution in June 2008 that stated — Be it resolved that ACET recommends that the Director of CDC fund and expedite implementation of currently available rapid drug-resistance assays in selected qualified reference labs to quickly identify drug-resistant TB, reduce transmission, and prevent further acquired drug resistance; such that by the end of 2008, labs are able to provide this assay for optimal patient care.
Introduction
In response to the ACET resolution, CDC convened an expert panel to examine the current status of rapid drug resistance testing in the United States, published evidence, and current guidelines and to provide guidance and make recommendations to CDC for developing a system to provide access to rapid drug-susceptibility testing to all TB Control programs in the United States. The panel included clinicians; control officials; laboratorians; and representatives from the TB Regional Training and Medical Consultation Centers, ACET, National TB Controllers Association, Association of Public Health Laboratories, and CDC. This report to the Director of the Division of TB Elimination describes general principles and considerations for molecular drug-resistance testing service from clinical, laboratory, and public health perspectives; possible scenarios for providing a molecular drug-resistance testing service; and recommendations for CDC.
To ensure access to state-of-the-art testing, the panel recommends that CDC establish regional laboratories to provide molecular drug-resistance testing services to state and local TB programs. The panel recommends that molecular drug-resistance testing be available for one AFB smear-positive or NAA-positive respiratory specimen or one M. tuberculosis culture from each TB patient or TB suspect (estimate testing 15,000 to 20,000 samples per year). A phased approach to developing and implementing a molecular drug-resistance testing service would be prudent. As an initial step, the expert panel strongly recommends that CDC immediately establish a service to provide molecular drug resistance testing for TB suspects and patients at high-risk of having MDR TB and those deemed high priority by the state or local TB program(estimate testing 2,500 samples per year). CDC is encouraged to explore using supplements to existing cooperative agreements to provide sufficient new funds to existing, proficient molecular drug-resistance testing laboratories to allow them to expand their capacities to meet this need. CDC, state TB programs, and partners should aggressively work towards establishing the protocols and procedures for (a) identifying patients for whom the testing would be of benefit, (b) submitting specimens to the molecular drug-resistance testing laboratories, (c) reporting results, and (d) additional testing to determine the susceptibility of rifampin-resistant samples to first-line and second-line anti-tuberculosis drugs.
For any approach, it will be essential to route requests for, and reports of, the molecular drug-resistance testing through the local or state TB control program because this would (a) provide early engagement of the TB Control Program in potential TB cases, (b) improve communications between TB clinicians, controllers, and laboratorians, (c) reinforce the important role played by state and local TB Programs and laboratories, (d) engage a person knowledgeable about molecular drug-resistance testing for TB early in the decision process, and (e) avoid excessive and inappropriate ordering of the molecular drug-resistance tests. The molecular drug-resistance testing services should be aligned or coordinated with the services of the TB Regional Training and Medical Consultation Centers to facilitate access to advice on the appropriate use and interpretation of molecular drug-resistance tests and on treatment of patients with drug-resistant TB. Another essential feature is that the detection of drug resistance must immediately trigger expedited (reflex) testing for susceptibility to first and second line anti-TB drugs by conventional culture-based methods and molecular genetic methods.
New funds will be needed for the molecular drug-resistance testing program. Funds in the current TB Elimination Cooperative Agreements should not be redirected to the molecular drug-resistance testing program.
Introduction
The emergence and spread of drug-resistant strains of Mycobacterium tuberculosis are greatly complicating tuberculosis (TB) control efforts in many countries. An estimated 511,000 cases of multidrug-resistant TB (MDR TB; caused by strains resistant to at least isoniazid and rifampin) occurred globally in 2007 (1). In the United States, a total of 125 cases of MDR TB were reported in 2007 (1.2% of culture-positive cases with susceptibility testing performed) as well as 2 cases of extensively drug resistant TB (XDR TB) (5). MDR TB is significantly more difficult and expensive to treat than drug-susceptible TB. It is estimated that preventing a single case of MDR TB would save the U.S. health care system more than $250,000 (4).
The management of drug-resistant TB cases starts with a reliable diagnosis, which is obtained by isolating M. tuberculosis bacteria from clinical specimens and conducting drug-susceptibility tests. The susceptibility of bacteria to a particular drug is usually determined by attempting to grow the bacteria in or on media containing that drug. The agar and liquid culture proportion methods are used by most laboratories in the United States that perform susceptibility testing of M. tuberculosis bacteria (6,7). Because of the slow growth of M. tuberculosis bacteria and the requirement for isolation before drug-susceptibility testing, the agar proportion method typically requires six to eight weeks to provide results while the liquid culture methods can provide results in four to five weeks. Molecular methods can reduce the time required for detection of drug resistance to one to two days. Because of the earlier detection of resistance and earlier initiation of effective therapy, the use of rapid molecular methods for detecting rifampin resistance may reduce periods of infectiousness of MDR TB cases by as much as six weeks, reduce the further spread of MDR TB, and improve treatment outcomes (2,3).
In recognition of the importance of rapid drug-susceptibility testing, a proposed revision of the Diagnostic Standards and Classification of Tuberculosis in Adults and Children (7) is likely to support the use of rapid molecular drug-resistance tests for AFB smear-positive sputum sediments from TB patients who are suspected to have drug-resistant disease or who are from a region or population with a high prevalence of drug resistance. In addition, the Advisory Council for the Elimination of Tuberculosis (ACET) passed a resolution in June 2008 that stated —
Be it resolved that ACET recommends that the Director of CDC fund and expedite implementation of currently available rapid drug-resistance assays in selected qualified reference labs to quickly identify drug-resistant TB, reduce transmission, and prevent further acquired drug resistance; such that by the end of 2008, labs are able to provide this assay for optimal patient care.
In response to the ACET resolution and the proposed revision of the diagnostic standards (7), CDC convened an expert panel to examine the current status of rapid drug-resistance testing in the United States, published evidence, and current guidelines and to provide guidance and make recommendations to CDC for developing a system to provide access to rapid drug-resistance testing. The expert panel included clinicians; control officials; laboratorians; and representatives from the ACET, TB Regional Training and Medical Consultation Centers (RTMCC), National TB Controllers Association (NTCA), Association of Public Health Laboratories (APHL), and CDC.
Background on molecular drug-resistance (DR) tests
Recent advances in the understanding of the molecular basis or genetics of drug resistance have enabled development of rapid, DNA-based, molecular tests to detect mutations associated with drug resistance. If a mutation thought to be associated with resistance is detected in such a rapid test, the bacteria are considered to be drug resistant. If no mutation is detected, the bacteria are assumed to be drug susceptible. The key advantage of the molecular tests is that they can provide results within 24 to 48 hours, because they take advantage of the speed of nucleic acid amplification. These tests have been referred to in various publications as genetic or molecular drug-susceptibility tests, genetic or molecular detection of drug resistance tests, molecular tests to detect drug (or antimicrobial or antibiotic)-resistance mutations, or tests to detect molecular or genetic markers of drug resistance. In this report, the tests will be referred to simply as molecular drug-resistance (DR) tests.
Mutations associated with resistance to many of the anti-TB drugs have been described (8,9). For example, ~95% of rifampin-resistant M. tuberculosis strains carry mutations within the rifampin resistance-determining region (RRDR), an 81-bp region of the rpoB gene. Because of the strong association between the presence of mutations in the RRDR and rifampin resistance, several molecular genetic tests to detect RRDR mutations have been developed and evaluated for their ability to detect resistance in clinical isolates. Genetic or molecular tests for detecting mutations are, in general, variations of nucleic acid amplification (NAA) tests. Typically, the polymerase chain reaction (PCR) is used to amplify a target sequence followed by a second assay to determine if the sequence contains a mutation associated with resistance, such as DNA sequencing or hybridization assays.
a. For hybridization assays such as the INNO-LiPA® Rif.TB (Innogenetics) and GenoType® MTBDR(plus) (Hain LifeScience GmbH) line-probe assays, the region of a gene associated with resistance is PCR amplified, and the labeled PCR products hybridized to oligonucleotide probes immobilized on a nitrocellulose strip. Mutations are detected by lack of binding to wild-type probes or by binding to probes specific for commonly occurring mutations. Compared to culture-based DS tests, the MTBDR(plus) line probe assay displays a pooled sensitivity of 0.98 and a pooled specificity of 0.99 for detecting rifampin resistance in isolates or directly from clinical specimens (10–12).
b. Molecular beacons are hybridization probes which emit fluorescence only when hybridized to their target and which can discriminate between targets differing only by a single nucleotide. In the California Microbial Diseases Laboratory, molecular beacon assays were designed to detect mutations in the rpoB gene directly from clinical specimens and from cultures. The results of rpoB molecular beacons tests showed 96% to 97% agreement with culture-based results in a series of ~1,000 clinical specimens and cultures (E. Desmond, personal communication).
c. Validation studies conducted at the Wadsworth Center of an approach that combines PCR-amplification of the entire 81 bp RRDR with pyrosequencing revealed that the test displayed a sensitivity of <1 colony forming unit, 100% specificity, and 99% agreement in the 188 cultures and specimens tested (13–15; K. Musser, personal communication).
Molecular DR tests for other anti-TB drugs are much less developed than the tests for rifampin resistance. A meta-analysis of the performance of the Hain MTBDR(plus) assay for detecting isoniazid resistance revealed a pooled sensitivity of 0.85 (95%CI 0.77– 0.90) and a pooled specificity of 0.99 (95%CI 0.98–1.00) (11,12). Validation studies conducted in the California Microbial Diseases Laboratory using archived cultures revealed that the molecular beacon test displayed 82.7% sensitivity, 100% specificity, 100% positive predictive value, and 98.1% negative predictive value for detecting isoniazid resistance (16, E. Desmond, personal communication).
The critical contribution of molecular DR tests for TB treatment and control is earlier detection of resistance: they can reliably detect mutations associated with drug resistance in 1 to 2 days. Not only does this reduce the time to detect rifampin resistance, but for MDR TB patients this also reduces the time from TB diagnosis to the start of MDR TB treatment and from the first positive culture to culture conversion by six weeks and improves patient outcomes (2,3). The reduction of the estimated infectious period after diagnosis by six weeks should have a large impact on public health measures to stop the spread of MDR TB.
General considerations and principles for a molecular drug-resistance testing service
1. 13,299 TB cases were reported to CDC in 2007 (5,17).
a. This includes 10,590 pulmonary and 2697 extrapulmonary cases, 10426 culture confirmed cases, 762 isoniazid-resistant cases, and 125 MDR TB cases.
b. Of the 10,590 pulmonary cases, 4864 were sputum smear positive for AFB, 4524 were smear negative and 7366 were sputum culture positive and 1878 were culture negative.
2. Rifampin resistance is a reliable surrogate (positive predictive value >95%) for MDR TB when isolated rifampin resistance is uncommon, as it is in the United States (18–20).
3. Molecular DR tests are useful for testing isolates and respiratory specimens directly. However, the currently available tests are highly reliable when used with AFB-smear positive specimens, but they are less reliable when used with AFB-smear negative specimens. There is little information on the performance of molecular DR tests with other types of specimens.
4. Both ‘susceptible’ and ‘resistant’ results from molecular DR tests can be useful.
a. The sensitivity and specificity of molecular DR tests for rifampin are sufficiently high (>97%) to use both resistant and susceptible results in case management decisions.
b. The sensitivity of molecular DR tests for isoniazid is not sufficient to exclude isoniazid resistance based on a negative result. However, because isoniazid resistance is about 8% in the United States, the positive predictive value for isoniazid resistance is relatively high and molecular detection of isoniazid resistance can be used in case management decisions.
5. Molecular DR testing is particularly useful for
a. patients suspected or at high risk of having drug-resistant TB,
b. very ill patients for whom drug-susceptibility information might alter case management decisions, such as patients who do not get better while taking standard first-line therapy,
c. outbreak or contact investigations when drug resistance is suspected in the source case or in some severely immunocompromised persons such as HIV-infected persons or those receiving dialysis in which knowledge of drug-susceptibility would be a significant benefit and affect preventive therapy decisions,
d. persons for whom drug-susceptibility information would influence TB Control decisions such as placing the person on a ‘Do Not Board’† list, and
e. isolates that contain a mixture of M. tuberculosis bacteria and other mycobacteria or respiratory specimens containing only nonviable M. tuberculosis bacteria.
6. Molecular DR testing has significant potential added value for clinicians and TB control officials.
a. Earlier detection of resistance leads to earlier initiation of an effective treatment regimen, a reduced period of infectiousness, and improved patient outcomes.
b. Earlier notification of drug-resistant TB cases should permit public health interventions sooner and may engage an MDR TB expert sooner in the care of the TB patient.
c. Earlier detection of rifampin resistance should lead to earlier testing for susceptibility to other first-line and second-line anti-TB drugs.
7. Benefits of routing requests for molecular DR tests through the TB Program include
a. early engagement of the TB Control Program in potential TB cases,
b. early engagement of TB laboratory in follow-up susceptibility testing,
c. improved communications between TB clinicians, controllers, and laboratorians,
d. reinforcement of the important role played by state and local TB Programs,
e. engagement of a person knowledgeable about molecular DR testing for TB early in decisions regarding whether or not to request a molecular DR test, and
f. avoidance of excessive and inappropriate ordering of the molecular DR tests – simply having a check-off box on a form often leads to inappropriate ordering.
8. Turnaround time (TAT) must be as brief as possible to maximize benefits of molecular DR testing. The key TAT is the interval from specimen collection to time that the test result is used by the clinician for case management.
9. It is essential that the detection of resistance immediately trigger additional (reflex) testing for susceptibility to first and second line anti-TB drugs by conventional and molecular methods.
10. A potential benefit of a system for providing molecular DR tests may be expanding access to NAA testing for the initial diagnosis of TB.
11. State regulations need to be addressed as part of developing a regional approach. For example, laboratories, regardless of location, that conduct testing for patients in New York must be certified by New York State.
12. No molecular DR test has been approved by the FDA for use in the United States, although well-characterized test kits are available in Europe and elsewhere. Several validated molecular DR tests (line-probe assays, molecular beacons, and DNA sequencing) based on analyte specific reagents, often called “home-brew” or “in-house” tests are used in the United States. Each test displays similar performance characteristics. At this time, data do not demonstrate clearly the superiority of one method over another.
a. Tests that detect M. tuberculosis DNA and drug resistance in one step have potential advantages related to lower cost, less hands-on time, simpler testing procedure, and use of a closed system. Although the currently available one-step systems have excellent performance with AFB-smear positive specimens and cultures, they perform less well when used with AFB-smear negative specimens.
b. A protocol that uses two methods (e.g., uses an optimized NAA assay to detect M. tuberculosis DNA (13–15) and a second assay to assess resistance) has potential advantages of increased sensitivity, particularly if AFB-smear negative specimens are tested, and the possibility of conducting more informative second tests (e.g., sequencing). Disadvantages may include a higher cost, more hands-on time, and potential end-product contamination.
13. Specimens suitable for molecular DR testing include cultures, processed specimens (sediments), and non-processed respiratory specimens. Because of the differences in the cost of testing non-processed (processing and molecular testing) and processed specimens (only molecular testing), programs must accurately project the anticipated numbers of non-processed and processed specimens to be tested to enable the molecular DR testing laboratories to estimate the cost of the services and CDC to adequately fund the service.
a. Advantages of processed specimens include (1) prior testing showed the sample was AFB+ and (2) it would not be necessary to process the specimen at the molecular DR laboratory which saves time and labor for the molecular DR testing laboratory. Potential disadvantages include possibly insufficient quantity of the remnant sample and possible errors introduced during processing may affect the molecular DR testing.
b. Advantages of non-processed specimens include (1) results with this specimen would be a check or confirmation of the results from conventional testing of another specimen, (2) a processing method optimized for molecular DR testing could be used, and (3) any errors in testing the first specimen would not affect the result of molecular DR testing. Potential disadvantages include (1) increased work load and cost for the molecular DR testing laboratory, (2) lengthening of the TAT at the molecular DR testing laboratory, (3) delays for obtaining a second specimen, and (4) sometimes a follow-up specimen may be AFB-negative due to sporadic shedding of TB bacilli.
c. Advantages of testing cultures using molecular DR test include increased sensitivity and accuracy of the molecular DR test. Potential disadvantages include the time needed to obtain an isolate and the expense of shipping viable cultures may be as much as 5-fold more than for specimens.
14. Molecular DR tests enhance but do not replace culture or conventional drug-susceptibility testing.
a. Molecular DR tests are not as sensitive as culture for detecting M. tuberculosis complex bacteria.
b. Molecular DR tests are not as sensitive as culture-based proportion tests for detecting resistant bacteria in a mixture of resistant and susceptible bacteria.
c. Molecular DR tests are most useful for rifampin, somewhat less useful for isoniazid, and not currently available for other anti-TB drugs although some are being developed.
d. False-positive and false-negative molecular DR test results do occur.
15. In the case of discrepancies between conventional and molecular DR test results for rifampin and isoniazid, clinicians should use the conventional DS results and clinical judgment for case management decisions until the discrepancy is resolved.
16. Failure of molecular DR tests can be caused by the presence of inhibitors in the sample that prevent or reduce NAA. Inhibitors appear to be present in 2% to 5% of respiratory specimens tested by NAA (21). Procedures must be in place to ensure that inhibition does not cause falsely negative or non-interpretable molecular DR test results. This may include internal controls to detect inhibitors and reflex repeat testing of samples suspected to contain inhibitors with steps taken to reduce inhibition (e.g., dilution or purification of DNA).
17. Shipping costs will be substantial. In the Florida molecular DR testing program, the contract cost of FedEx next-day shipments for specimens is $11 to $28 ($3 to $5 for shipping container plus $8 [weekday] or $23 [Saturday] for transport). The cost of U.S. Postal Service next day shipments of specimens is $16.75 plus the cost of the container. For isolates, the cost of FedEx is $97 to $115 ($12 to $15 for the container plus $85 [weekday] or $100 [Saturday] for transport). The cost of shipping isolates to the genotyping laboratories using the CDC FedEx account is $27 per shipment plus the cost of the container.
18. Reagent costs for the currently validated molecular DR tests range from $8 to $30 per sample. It is estimated that one technician can perform 20 molecular DR tests per day. Additional operating costs include time and materials needed for processing samples; preparing samples for molecular DR testing; entering, verifying and reporting results; technical assistance and consultation; proficiency testing; quality laboratory management; equipment; information technology, and overhead.
19. Cost efficiency, rapid turnaround time, and expertise would be enhanced by establishing high-volume regional laboratories offering molecular DR tests.
20. New funds will be needed to cover the costs of the molecular DR testing program. Potential sources of non-CDC funds to partially offset the cost of the program include the Robert Wood Johnson Foundation, Centers for Medicare and Medicaid Services, and insurance providers.
21. Good communication between laboratorians, clinicians, and public health officials will be critical to optimizing the benefits of molecular DR testing. Standard language or statements to include in laboratory reports of molecular DR test results are needed, such as the information in points 4a, 4b, and 16 above, to assist clinicians interpret the results.
22. Education of laboratorians, clinicians, TB controllers, and policy makers on the appropriate use and interpretation of molecular DR tests for TB will be essential.
Possible scenarios and scope of testing for a molecular DR testing service
A phased approach to developing and implementing a molecular DR testing service would be prudent. In the initial phase, testing might be offered for TB patients or suspects at high-risk of having MDR TB and situations deemed high priority by the program (judicious use testing). A long-range goal should be to offer testing for all TB patients and suspects (universal testing). In addition to molecular DR tests, the resources of the molecular DR laboratory might be leveraged to provide other services for state and local TB programs and laboratories. The scope of any additional service, such as NAA testing for detection or culturing or second-line drug susceptibility testing, must be clearly defined and adequately funded.
Judicious use testing would concentrate on testing (a) samples for which the test result would alter case management or TB Control decisions, outbreak or contact investigations, preventive therapy in immunocompromised contacts, infection control, or Do Not Board lists; (b) samples from persons at risk of having drug-resistant TB (persons exposed to an MDR-TB case, from a population with a high rate of MDR TB, or failing or having failed therapy with first-line anti-TB drugs); and (c) respiratory specimens or isolates that can not be tested easily with conventional methods (non-viable specimens; mixed or contaminated cultures). Given that there are 100 to 150 new cases of MDR-TB reported to CDC each year and many new TB cases are persons from populations with a high prevalence of MDR TB, one would estimate that a judicious use molecular DR testing program would entail the testing of about 2500 samples per year. One or two regional molecular DR testing laboratories would be needed. The estimated cost of this is $300,000 to $400,000 plus the cost of shipping (~$70,000) and initial equipment.
Universal testing would involve molecular DR testing one AFB smear-positive or NAA-positive respiratory specimen or one M. tuberculosis culture from each TB patient or TB suspect. About 5000 AFB-smear positive pulmonary TB cases were reported to CDC in 2007. An approximately equal number of patients were AFB-smear positive due to the presence of non-tuberculous mycobacteria (NTM) in the respiratory specimen. About 7,400 pulmonary and ~3000 extrapulmonary culture-confirmed TB cases were reported to CDC in 2007. Thus, universal testing would entail testing 10,000 to 20,000 samples per year. Up to four regional molecular DR testing laboratories would be needed to handle this work load. The estimated cost of a universal molecular DR testing service is 1.2 million to $2 million dollars plus the cost of shipping ($250,000 to $500,000) and initial equipment.
Variations of the molecular DR testing options described above may allow CDC to address the needs of programs for NAA testing for detection as well as molecular DR testing. However, linking NAA testing for detection with molecular DR testing must be carefully thought through to determine if it is a cost-effective, reliable approach to providing molecular DR testing services to state and local TB programs.
1. Option 1: only samples shown to be NAA-positive for TB would be accepted by the molecular DR testing laboratory. In this case, universal testing would involve 7000 to 9000 samples because NAA tests detect 70% to 90% of pulmonary TB cases that are ultimately culture confirmed. This approach (a) would delay sample submission to the molecular DR testing laboratory by 1–2 days, although a positive NAA result at the local laboratory might prompt earlier initiation of therapy; (b) would increase the cost of the molecular DR testing service to the TB program to include the cost of NAA testing at the local laboratory; (c) might complicate the submission process for private- and public-sector laboratories and programs that do not have access to NAA testing, although this requirement might be an incentive for local laboratories to offer NAA testing; and (d) might reduce shipping costs if leftover DNA from the NAA testing were shipped. If NAA testing were required prior to submission, a phased implementation of this requirement would be essential to ensure that all programs have access to molecular DR testing when needed, perhaps by allowing programs to submit samples from patients meeting the judicious use criteria.
2. Option 2: the molecular DR testing laboratory would conduct NAA testing for detection as well as molecular DR testing. For AFB-smear positive specimens, the available molecular DR tests can reliably detect M. tuberculosis DNA, so a separate test for detection is not needed. For AFB-smear negative samples, an optimized NAA test for detection could be coupled with a molecular DR test to increase reliability of the molecular DR test. However, the performance of molecular DR tests with AFB-smear negative, NAA positive specimens is not known. This approach would (a) provide access to NAA testing for detection to local and state TB programs; (b) increase the cost of the molecular DR testing service to include the cost of NAA tests for detection; (c) allow use of a specimen processing method optimized for molecular DR testing; and (d) require strict criteria for submitting AFB-smear negative specimens to avoid inappropriate ordering of NAA tests for patients who are unlikely to have TB.
For any of the scenarios, a phased approach would be prudent. At a minimum, it would be essential to provide molecular DR testing services for TB patients or suspects at high-risk of having MDR TB and those deemed high priority by the program. This could be accomplished by providing sufficient new funding to existing, proficient molecular DR testing laboratories to expand their capacities to meet this need. If done through supplements to existing cooperative agreements, this might be done quickly. Such an interim service could serve as pilot projects and would allow time to (a) compare the performances and costs of currently available tests and select one or more for use in the molecular DR testing service; (b) assess and overcome potential obstacles and barriers to a regional approach to diagnostic testing such as local regulations regarding out-of-state testing, reporting requirement, and need for memoranda of agreement; (c) develop a strategy to coordinate or integrate services provided by the molecular DR testing and genotyping laboratories to avoid unnecessary duplication of efforts and shipment of isolates; (d) develop a strategy for implementing the molecular DR testing service to include informing potential service users of the availability of the service, how to access the service, and the appropriate use and interpretation of molecular DR tests for TB; (e) design a molecular DR testing service to meet the needs of local and state TB control programs; and (f) develop, compete, and award a contract to provide the services.
Research needs
1. Conduct operational, translational, and implementation research for developing, evaluating, and selecting the most effective testing algorithms for routine use and specific scenarios.
2. Evaluate the cost and benefits of molecular DR testing.
3. Evaluate and compare the performance of currently available tests to facilitate the choice of test(s) to use in the molecular DR testing service.
4. Develop and evaluate optimal specimen collection, transport, and processing methods.
5. Characterize the performance of molecular DR tests with mixtures of M. tuberculosis and NTM, mixtures of resistant and susceptible bacteria, different types of specimens, and cultures.
6. Characterize the performance of molecular DR tests with AFB-smear negative, NAA positive respiratory specimens.
7. Define the molecular basis of resistance to each first-line and second-line anti-TB drug.
8. Develop and evaluate molecular DR tests for first-line and second line anti-TB drugs. Tests are needed for drugs for which conventional testing is problematic (e.g., ethambutol, pyrazinamide) and the XDR TB defining drugs particularly the fluoroquinolones.
9. Conduct regulatory quality trials for molecular DR tests aimed at obtaining FDA approval.
10. Determine the value of the detection of individual mutations for predicting clinically significant drug resistance.
General Recommendations of the Expert Panel
1. All U.S. clinicians and public health TB programs should have access to molecular DR tests to aid in the diagnosis, treatment, and control of TB.
2. Molecular DR testing should be performed on one AFB smear-positive or NAA-positive respiratory specimen or one M. tuberculosis culture from each TB patient or TB suspect.
a. Testing should also include specimens regardless of AFB smear result or isolates from persons that the TB Control Program designates as high priority for molecular DR testing. However, programs must be aware that the performance of molecular DR tests with AFB-smear negative specimens has not been established.
b. Testing of a second sample (specimen or isolate) from a patient would be appropriate in situations deemed high priority by the TB program (e.g., a patient who is failing first-line therapy even though the initial molecular DR test indicated rifampin susceptibility or relapse in a patient who was non-adherent to the initial treatment plan).
3. A phased approach to developing and implementing a molecular DR testing service is recommended. For example, the initial service could provide molecular DR testing for TB patients or suspects deemed high priority by the TB program, while CDC and partners design and implement a feasible, practical, universal molecular DR testing service.
4. State and local TB control programs should develop, disseminate, and implement a protocol that enables health care providers in their jurisdiction to access the regional molecular DR testing services, including specifying criteria for selecting TB suspects or patients for testing. A standard test request (sample submission) form should be developed.
5. The initial molecular DR testing service should include detection of mutations associated with rifampin resistance and those associated with isoniazid resistance. The molecular DR testing service should incorporate molecular DR tests for fluoroquinolones, aminoglycosides, and drugs for which conventional testing is problematic (e.g., ethambutol, pyrazinamide) as they are validated.
6. Because data are not available at this time that clearly demonstrate the superiority of any of the currently validated methods over another, the panel does not recommend which test should be used in the molecular DR testing service. The decision of which test to implement in the molecular DR testing service may ultimately rest upon cost, performance, throughput, and turnaround time. Whichever technology is used, validation of the test and meeting all pertinent CLIA and FDA regulations by the molecular DR testing laboratory are essential.
7. The molecular DR testing service should be designed such that it is able to take advantage of improvements in technologies and the understanding of the molecular basis of drug resistance.
8. The possibility of linking NAA testing for detection of M. tuberculosis with molecular DR testing either sequentially (local laboratory to molecular DR testing laboratory) or as a combined test at the molecular DR testing laboratory should be explored to determine if it would be is a cost-effective, reliable approach to providing services to state and local TB programs.
9. Up to four laboratories will be needed initially to provide universal molecular DR testing and their services should be coordinated with the services of the TB RTMCCs. This would provide (a) increased molecular DR testing capacity, (b) a reasonable workload per laboratory which may facilitate meeting turnaround times, (c) redundancy and surge capacity, (d) geographic distribution, (e) close collaboration with experts in the treatment of MDR TB, and (f) opportunities for rechecking and external quality control.
10. Molecular DR testing laboratories should have the ability to test isolates and processed and non-processed specimens. Only respiratory specimens should be routinely tested. Other specimens such as CSF or tissue samples may be tested in priority situations.
a. In a rollout phase, the molecular DR testing laboratories might primarily test processed specimens. In this phase, specimens would be collected, sent to the state or local public health laboratory, processed at the state or local laboratory, determined to be AFB-positive, and submitted to the molecular DR testing laboratory.
b. The numbers of non-processed and processed specimens tested will depend on the protocols developed by local programs to select patients and submit specimens. TB programs must provide reliable estimates of the numbers of non-processed and processed specimens to be submitted to enable the molecular DR testing laboratories to project the costs of molecular DR testing,
11. The interval from specimen collection to reporting of the test result to the treating clinician must be as brief as possible. Laboratories and programs should track this performance measure.
a. Specimens must be delivered promptly to the molecular DR testing laboratory.
i. An overnight delivery service should be used. State programs may need to provide training for local laboratorians in packaging and shipping, because delivery services such as FedEx only accept shipments packaged by a certified shipper.
ii. Laboratories must promptly package and ship samples to the molecular DR testing laboratory and avoid delays associated with batching specimens for shipment. For non-processed specimens, this is probably the day the specimen is collected. For processed specimens, this is probably the day after the specimen is received in the primary laboratory.
b. Specimens must be tested promptly in the molecular DR testing laboratory, preferably on the day received (i.e., without introducing significant delays by batching specimens).
c. Six day a week service is preferred.
d. The molecular DR test results should be available within 2 business days of specimen receipt.
e. An initial positive molecular DR test result must be treated as a critical test value. It must be immediately reported to the clinician and to public health authorities. Laboratorians should be available for consultation as to test interpretation and need for follow-up testing.
12. Detection of rifampin resistance must trigger expedited, reflex testing for susceptibility to first-line and second-line drugs (SLD) by conventional culture-based methods and available molecular methods. This could be done at the molecular DR testing laboratory, the submitting laboratory, a state public health laboratory, a center of excellence for SLD testing, or CDC.
13. Each TB program should designate who would be notified of the molecular DR test results.
a. The preferred method of reporting is via electronic means such as secure email or posting results on a secure web site.
b. The detection of drug resistance in specimen or isolate should be reported by telephone to facilitate prompt action by the program and clinician.
c. Standardized reporting language should be developed and used.
d. In all cases, reporting must meet requirements for maintaining patient confidentiality.
14. Procedures for detecting and reporting discrepancies between the results of molecular and conventional testing must be developed and implemented. Clinicians should use clinical judgment and the conventional DS result for isoniazid and rifampin for case management decisions, until the discrepancy is resolved.
a. The responsibility for identifying discrepancies lies with those having timely access to the molecular DR and conventional DS results. This may be the treating clinician, TB program or public health laboratory.
b. Procedures must be in place for reporting discrepant results and providing consultation.
c. Regardless of who detects a discrepancy, protocols are needed for distributing information to all involved parties and follow-up testing to resolve the discrepancy.
15. Protocols for analyzing discrepancies between conventional and molecular DR results must be developed and implemented.
a. The molecular DR test should be repeated on the remnant of the original specimen or a sample of the culture from the patient.
b. The initial molecular DR result should be evaluated for concerns such as unusual amplification or evidence of a mixed population of bacteria (a low percentage of resistant bacteria may lead to false-susceptible molecular DR results).
c. The conventional DS result should be evaluated for concerns such as contamination that might produce false-resistant results.
d. Repeating the conventional DS tests should be considered.
e. A sample of the culture patient should be submitted to a referee laboratory (e.g., CDC) for additional molecular (e.g., sequencing) and conventional testing.
16. State and local TB programs should share some of the cost of the molecular DR testing. One possibility would be for programs or test requestors to pay the cost of shipping specimens to the molecular DR testing laboratories.
17. The activities of the molecular DR testing and TB genotyping laboratories should be coordinated (possibly integrated) to avoid unnecessary duplication of efforts at the local or regional laboratory for shipping and testing of isolates.
18. A reliable laboratory service includes procedures for internal and external quality control and a robust monitoring and evaluation plan.
Communication Plan for the Report
The panel report recommends disseminating the report, which is provided to the Director of the Division of TB Elimination, by posting on the CDC website and direct distribution to key stakeholders in order to reach clinicians, TB control officials, laboratorians, governmental organizations, regulatory agencies, policy makers, and other TB partners.
Recommendations
1. CDC should develop a system with sufficient testing capacity to enable molecular DR testing for one AFB smear-positive or NAA-positive respiratory specimen or one M. tuberculosis culture from each TB patient or TB suspect and specimens or isolates from persons that the local or state TB Control Program designates as high priority for testing.
2. CDC should evaluate existing molecular DR testing services to identify best practices.
3. CDC should use a phased approach to implementing a universal molecular DR testing service.
4. CDC should immediately establish an interim service to provide molecular DR testing for persons at high-risk of having MDR TB and those deemed high priority by the local TB program. CDC is encouraged to explore using supplements to existing cooperative agreements to provide sufficient new funds to existing, proficient molecular DR testing laboratories to allow them to expand their capacities to meet this need. The interim service could serve as a pilot project to inform the development of a universal molecular DR testing service.
5. CDC should establish and fund regional laboratories to provide molecular DR testing for state and local TB programs. Funds in the current TB Elimination Cooperative Agreements should not be redirected to the molecular DR testing program. The molecular DR testing laboratories should
a. coordinate molecular DR testing services with the medical consultation and training services of the TB Regional Training and Medical Consultation Centers (RTMCCs),
b. provide six-day-a-week service,
c. use validated molecular methods to detect rifampin and isoniazid resistance,
d. implement molecular DR testing for anti-TB drugs other than rifampin and isoniazid (e.g., fluoroquinolones) as the tests are developed and validated,
e. report results electronically within two business days of specimen receipt,
f. report detection of drug resistance in specimen or isolate by telephone to facilitate prompt action by the program and clinician,
g. ensure notification of appropriate individuals (e.g., local program, laboratory, clinician) of the need for expedited testing of rifampin-resistant samples for susceptibility to first-line and second-line anti-TB drugs, and
h. participate in an external quality assurance program.
6. CDC should work with TB partners and state and local TB programs and laboratories to identify and overcome potential obstacles and barriers to implementing a regional molecular DR testing service, such as local regulations regarding out-of-state testing, certification of laboratories, reporting requirements, and need for memoranda of agreement.
7. CDC and partners should develop clear policies and standard operating procedures for referring specimens to the molecular DR testing laboratories.
a. CDC should develop and fund a process for shipping M. tuberculosis cultures to the molecular DR testing laboratories.
b. CDC should develop and fund a process for shipping specimens to the molecular DR testing laboratories for TB laboratories or programs that can not afford the cost of shipping.
8. CDC should coordinate, and possibly integrate, activities of the molecular DR testing and genotyping laboratories to avoid unnecessary duplication of efforts and shipment of isolates.
9. CDC should work with partners to develop external quality assurance, proficiency testing, and rechecking programs for the molecular DR testing service.
10. CDC should develop a robust process for monitoring and evaluating the performance of the molecular DR testing laboratories. This should include post-market surveillance to determine the performance, cost, and benefit of the molecular DR tests as performed in a regional testing service.
11. CDC should work with partners to develop protocols to analyze discrepancies in the results of molecular DR and conventional tests. CDC should collect data on and investigate discrepancies to better understand the performance of molecular and conventional DS testing.
12. CDC, NTCA, and APHL should convene a work group to develop guidelines, templates, and models for programs to use in developing their systems to access the molecular DR testing service and receive reports.
13. CDC should work with partners such as APHL and NTCA to assess training needs, develop training materials, and establish an education program for TB control officials, laboratorians, clinicians, and policy makers on the appropriate use and interpretation of molecular DR tests for TB.
14. CDC should work with partners such as APHL and NTCA to develop a process for providing guidance, technical assistance, and consultation on clinical, programmatic, and laboratory aspects of the appropriate use and interpretation of molecular DR tests for TB in the United States.
15. CDC should develop a broader evidence base to support changes in recommendations and practices and investigate the economic implications of molecular DR testing.
16. CDC should develop and promote a research agenda for molecular DR testing for TB.
17. CDC should work with private- and public-sector partners to increase the number and types of molecular DR tests, commercial sources, FDA-approved tests, and validated tests.
a. CDC and FDA should encourage manufacturers to develop molecular DR tests for TB and submit to FDA for review and approval.
b. CDC should assist manufacturers with regulatory quality trials of molecular DR tests aimed at receiving FDA approval.
c. CDC should establish a repository of well-characterized isolates for use in developing, evaluating, and validating molecular DR tests for TB.
18. CDC should disseminate the panel report and any resulting CDC recommendations in multiple media, in order to reach clinicians, TB control officials, laboratorians, regulatory agencies, policy makers, and other TB partners. This may include publication in scientific or medical journals or MMWR, posting on the CDC website, use of electronic mail lists, and direct distribution to key stakeholders.
19. CDC should monitor and evaluate the implementation of the recommendations. CDC should periodically, perhaps annually, provide progress reports to ACET.
References
1. World Health Organization. The WHO/IUATLD Global Project on Anti-Tuberculosis Drug Resistance Surveillance. Antituberculosis Drug Resistance in the World Report No. 4 Vol. WHO/HTM/TB2008.394. Geneva, Switzerland, 2008.
2. O'Riordan P, Schwab U, Logan S, Cooke G, Wilkinson RJ, Davidson RN, Bassett P, Wall R, Pasvol G, Flanagan KL. Rapid molecular detection of rifampicin resistance facilitates early diagnosis and treatment of multi-drug resistant tuberculosis: case control study. PLoS ONE. 2008;3:e3173.
3. Allen J, Lin G, Desmond E, Westenhouse J, Mase S, Scott C, Schecter G, Flood J. Can rapid drug resistance testing improve time to effective treatment start for multidrug-resistant tuberculosis cases? 5th National Conference on the Laboratory Aspects of Tuberculosis. San Diego CA. 2008.
4. Farmer P and Kim JY. Community based approaches to the control of multidrug resistant tuberculosis: Introducing DOTS-plus. Brit Med J. 2008;317:671–674.
5. CDC. Trends in Tuberculosis — United States, 200. MMWR 2009;58:249-53.
6. NCCLS., Susceptibility Testing of Mycobacteria, Norcardiae, and Other Aerobic Actinomycetes; Approved Standard M24-A. 2003, Wayne, Pennsylvania.
7. American Thoracic Society; CDC; Council of the Infectious Disease Society of America. Diagnostic standards and classification of tuberculosis in adults and children. Am J Respir Crit Care Med 2000;161:1376–95.
8. Zhang Y and Telenti A. Genetics of drug resistance in Mycobacterium tuberculosis. In Hatfull GF and Jacobs WR Jr (Eds.). Molecular Genetics of Mycobacteria. pp. 235–54. ASM Press, Washington, D.C. 2000.
9. Johnson R, Streicher EM, Louw GE, Warren RM, van Helden PD, Victor TC. Drug resistance in Mycobacterium tuberculosis. Curr Issues Mol Biol. 2006;8:97-111.
10. Morgan M, Kalantri S, Flores L, Pai M. A commercial line probe assay for the rapid detection of rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. BMC Infect Dis. 2005;5:62.
11. Ling DI, Zwerling AA, Pai M. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur Respir J. 2008;32:1165–74.
12. WHO Expert Group Report. Molecular line probe assays for rapid screening of patients at risk of multi-drug resistant tuberculosis (MDR-TB). Geneva: World Health Organization; 2008. http://www.who.int/tb/features_archive/expert_group_report_june08.pdf.
13. Halse TA, Cunningham P, Wolfgang WJ, Dumas N, and Musser KA. Development and implementation of a real-time PCR assay for rapid identification of Mycobacterium tuberculosis complex DNA from clinical samples in New York State. American Society for Microbiology 108th General Meeting. Boston, MA. 2008.
14. Cunningham PL, Driscoll J, Rivenburg J, McGarry M, Halse TA, Musser KA, and Escuyer VE. Implementation of a real-time PCR assay for the rapid identification of Mycobacterium tuberculosis complex DNA from clinical samples in New York State. 5th National Conference on Laboratory Aspects of Tuberculosis. San Diego, CA. 2008.
15. Halse TA, Edwards J, Driscoll JR, Escuyer VE, and Musser KA. A pyrosequencing approach to rapidly assess mutations in the rpoB gene associated with rifampin resistance in clinical specimens of Mycobacterium tuberculosis. 48th Annual ICAAC/IDSA. Washington, DC. 2008.
16. Lin S-YG, Lin S-Y, Probert W, Lo M , Desmond E. Rapid detection of isoniazid and rifampin resistance mutations in Mycobacterium tuberculosis complex from cultures or smear-positive sputa by use of molecular beacons. J Clin Microbiol 2004;42:4204-08.
17. CDC. Reported tuberculosis in the United States, 2007. Atlanta, GA: U.S. Department of Health and Human Services, CDC, September 2008.
18. Traore H, Fissette K, Bastian I, Devleeschouwer M, Portaels F. Detection of rifampicin resistance in Mycobacterium tuberculosis isolates from diverse countries by a commercial line probe assay as initial indicator of multidrug resistance. Int J Tuberc Lung Dis. 2000;4:481-4.
19. LoBue PA, Moser KS. Isoniazid- and rifampin-resistant tuberculosis in San Diego County, California, United States, 1993-2002. Int J Tuberc Lung Dis. 2005;9:501-6.
20. Moore M, Onorato IM, McCray E, Castro KG. Trends in drug-resistant tuberculosis in the United States, 1993-1996. JAMA. 1997;278:833-7.
21. Dinnes J, Deeks J, Kunst H, Gibson A, Cummins E, Waugh N, Drobniewski F, Lalvani A. A systematic review of rapid diagnostic tests for the detection of tuberculosis infection. Health Technol Assess 2007;11(3):1–196.
Panel Members
Edward P. Desmond, PhD, Chief, Mycobacteriology and Mycology Section, Microbial Diseases Laboratory, California Dept. of Public Health
Kimberly Field, RN, MSN, Director, TB Control Services, Washington Department of Health
Phil Griffin, BBA, Director, Kansas TB Control and Prevention, Kansas Department of Health and Environment
Denise Ingman, BS, MT DPHHS, Project Director, Montana Department of Public Health and Human Services
Kimberlee A. Musser, PhD, Director of Bacteriology, Wadsworth Center, New York State Department of Health
Margaret J. Oxtoby, MD, Director, Bureau of Tuberculosis Control, New York State Department of Health
Virginia A. Pruitt, Acting Director, Respiratory Diseases Division, Bureau of Clinical Laboratories, Alabama Department of Public Health
Jafar H. Razeq, PhD, Division Chief, Public Health Microbiology Laboratories Administration, Maryland Department of Health
Max Salfinger, MD, Chief, Bureau of Laboratories, Florida Department of Health
Barbara Seaworth, MD, Medical Director, Heartland National TB Training and Education Center
James P. Watt, MD, MPH, Chief, Tuberculosis Control Branch, California Department of Public Health
Kelly E. Wroblewski, MPH, MT (ASCP), HIV, STD, TB, Hepatitis Program Manager, Association of Public Health Laboratories
Suzanne Zanto, CLS(NCA) SM(NRM), Microbiology and Molecular Laboratory Manager, Montana Public Health Laboratory
Division of TB Elimination, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention
Kashef Ijaz MD, Chief, Field Services and Evaluation Branch
Beverly Metchock DrPH, Chief, Reference Laboratory Team, Mycobacteriology Laboratory Branch
James Posey PhD, Chief, Applied Research Team, Mycobacteriology Laboratory Branch
Thomas M. Shinnick PhD, Associate Director for Global Laboratory Activities.
Angela Starks PhD, Mycobacteriology Laboratory Branch
Appendix 1: Molecular Basis of Drug Resistance and Molecular DR tests
Drug resistance in Mycobacterium tuberculosis bacteria arises mainly through the acquisition of mutations in the chromosomal sequence that encode changes that 1) block the activity of a drug (mutations in rpoB prevent binding of rifampin to RNA polymerase and inhibition of transcription), 2) block activation of a prodrug (e.g., mutations in katG lead to loss of the ability of catalase to activate the prodrug isoniazid to its active form), or 3) produce an activity that binds or destroys the drug (e.g., mutations in inhA increase the amount of InhA protein which interferes with the activity of isoniazid by binding sufficient isoniazid to reduce its effective concentration in the bacterium to below an inhibitory level) (1,2). The mutations associated with resistance to many of the antituberculosis drugs have been identified, though much work remains to be done to identify the molecular basis of resistance for some of the drugs and to determine the predictive value of finding a particular mutation in a strain of M. tuberculosis (1,2). For example, approximately 95% of rifampin-resistant M. tuberculosis strains carry mutations within the rifampin-resistance determining region (RRDR), an 81-bp region encoding codons 507 through 533 of the rpoB gene.
Molecular genetic tests for detecting drug-resistance are, in general, just a variation of nucleic acid amplification (NAA) tests and can reliably provide information on the presence of mutations associated with drug resistance in 1 to 2 days. Typically, PCR is used to amplify a target sequence followed by a second assay to determine if the sequence contains a mutation associated with resistance. Methods that have been described for the latter include DNA sequencing, pyrosequencing, electrophoretic detection methods (e.g., single strand conformation polymorphism), methods for detecting mismatches in heteroduplexes (e.g., temperature gradient HPLC analysis or branch migration inhibition), and hybridization assays (e.g., molecular beacons, microarrays, membrane hybridization, or line-probe assays). Kits for detecting mutations associated with rifampin resistance that are commercially available in Europe and elsewhere include line-probe assays (INNO-LiPA® Rif.TB, Innogenetics and GenoType® MTBDR(plus), Hain LifeScience GmbH) and microarray assays (CombiChip Mycobacteria DR, GENE IN). Some also detect mutations associated with isoniazid resistance. In-house PCR-based tests using molecular beacons have also been used for diagnostic purposes in a few clinical laboratories.
For the hybridization assays, the region of the target gene associated with resistance is PCR amplified, and the labeled PCR products hybridized to oligonucleotide probes immobilized on a nitrocellulose strip or in a microarray. Mutations are detected by lack of binding to wild-type probes and/or by binding to probes specific for commonly occurring mutations. The performance of the line-probe assays relative to culture-based DS tests was evaluated in meta-analyses (3–5). For the INNO-LiPA Rif.TB assay, the pooled sensitivity was 0.97 (95%CI 0.95–0.98) and the pooled specificity was 0.99 (95%CI 0.98–1.00) for detecting rifampin resistance in M. tuberculosis isolates. Overall discriminatory ability of the assay was 99% and overall accuracy was 97%, with all studies yielding consistently high performances. In four studies, the INNO-LiPA Rif.TB showed 100% specificity and sensitivity ranging from 80% to 100% for detecting rifampin resistance directly from clinical specimens. For the MTBDR and MTBDR(plus) assays, the pooled sensitivity was 0.98 (95%CI 0.96–0.99) and the pooled specificity was 0.99 (95% CI 0.97–0.99) for detecting rifampin resistance in isolates or directly from clinical specimens. Overall discriminatory ability of the assay was 99% and overall accuracy 97%, with all studies yielding consistently high performances.
Molecular beacons are hybridization probes which emit fluorescence only when hybridized to their target. Molecular beacons can discriminate between targets differing by a single nucleotide. Because molecular beacons can use different fluorophores, real-time PCR assays can be designed in which different DNA fragments or mutations can be amplified and detected simultaneously in the same tube. For example, a single-well assay has been developed that uses five molecular beacons to detect mutations associated with rifampin resistance in M. tuberculosis bacteria and appears to perform similarly as the line-probe assays. In the California Microbial Diseases Laboratory, molecular beacons were designed to detect mutations in rpoB, katG, and inhA promoter region genes and directly applied to clinical specimens or to cultures. Comparison of molecular beacons results with results of culture-based drug-susceptibility testing showed 96% to 97% agreement in a series of approximately 1,000 clinical specimens and cultures (6, E. Desmond, personal communication).
Validation studies were conducted at the Wadsworth Center of an approach that combines PCR-amplification of the RRDR with rapid (< 2hrs) DNA sequencing (K. Musser, personal communication). A PyrosequencingTM protocol utilizing two primers was developed to sequence the 81-bp RRDR of the rpoB gene and obtain a clear and accurate pyrogram. The detection limit was determined and the pyrosequencing approach was evaluated in primary specimens positive for M. tuberculosis complex DNA by real-time PCR. Final results were compared with conventional susceptibility testing results and/or DNA sequencing. This test has a detection limit of <1 colony forming unit, 100% specificity, and 99% agreement in the 188 cultures and specimens tested. (7)
Molecular genetic tests for the other antituberculosis drugs are much less developed and studied than the tests for rifampin resistance. A meta-analysis of the performance of the Hain MTBDR(plus) assay for detecting isoniazid revealed a pooled sensitivity of 0.85 (95%CI 0.77– 0.90) which ranged from 57%–100% and a pooled specificity of 0.99 (95%CI 0.98–1.00) which was fairly consistent across studies. Validation studies conducted in the California Microbial Diseases Laboratory that used archived cultures revealed that the molecular beacon test displayed 82.7% sensitivity, 100% specificity, 100% positive predictive value, and 98.1% negative predictive value for detecting isoniazid resistance (6). Tests for the other key resistances, especially the XDR TB defining resistances, are in various stages of development from discovery of the mutations associated with resistance to development of prototype assays and laboratory-based evaluations.
References
1. Zhang Y and Telenti A. (2000) Genetics of drug resistance in Mycobacterium tuberculosis. In Hatfull GF and Jacobs WR Jr (Eds.). Molecular genetics of Mycobacteria (ASM Press, Washington, D.C) pp. 235–54.
2. Johnson R, Streicher EM, Louw GE, Warren RM, van Helden PD, Victor TC. Drug resistance in Mycobacterium tuberculosis. Curr Issues Mol Biol. 2006 Jul;8(2):97-111.
3. Morgan M, Kalantri S, Flores L, Pai M. A commercial line probe assay for the rapid detection of rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. BMC Infect Dis. 2005; 5:62.
4. Ling DI, Zwerling AA, Pai M. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur Respir J. 2008;32:1165–74.
5. WHO Expert Group Report. Molecular line probe assays for rapid screening of patients at risk of multi-drug resistant tuberculosis (MDR-TB). Geneva: World Health Organization; 2008. http://www.who.int/tb/features_archive/expert_group_report_june08.pdf.
6. Lin S-YG, Lin S-Y, Probert W, Lo M , Desmond E. Rapid detection of isoniazid and rifampin resistance mutations in Mycobacterium tuberculosis complex from cultures or smear-positive sputa by use of molecular beacons . J Clin Microbiol 2004 42: 4204-08.
7. Halse TA, Edwards J, Driscoll JR, Escuyer VE, and Musser KA. A pyrosequencing approach to rapidly assess mutations in the rpoB gene associated with rifampin resistance in clinical specimens of Mycobacterium tuberculosis. 48th Annual ICAAC/IDSA Annual Meeting. Washington, DC. 2008.
Appendix 2: Roles and Responsibilities in a Regional Laboratory System for Providing for Molecular Drug-Resistance Testing for TB
CDC should establish regional laboratories to provide molecular DR testing for rifampin resistance and isoniazid resistance for TB control programs in the United States and affiliated jurisdictions. The goal is to provide sufficient testing capacity to enable molecular drug-resistance testing for (1) one AFB smear-positive or NAA-positive respiratory specimen or one Mycobacterium tuberculosis culture from each TB patient or TB suspect (‘universal testing’) and (2) specimens or isolates from persons that the TB Program designates as high priority for molecular DR testing (‘judicious use testing’). For universal testing, it should be noted that there were ~5000 pulmonary AFB-smear-positive TB cases reported to CDC in 2007; an approximately equal number of AFB-smear-positive specimens due to non-tuberculosis mycobacteria; and ~7,400 pulmonary and ~3000 extrapulmonary culture-confirmed TB cases reported to CDC in 2007. Thus, universal testing would entail testing 10,000 to 20,000 samples per year.
A phased approach to developing and implementing a molecular DR testing service will be needed. As a first step, it would be essential to provide molecular DR testing services for testing TB patients or suspects at high-risk of having MDR TB and those deemed high priority by the program. This could be accomplished by providing sufficient new funding to existing, proficient molecular DR testing laboratories to allow them to expand their capacities to meet this need. If done as supplements to existing cooperative agreements, this could be done quickly. These programs could also serve as pilot projects by offering universal testing for selected programs. Such an interim service would allow time to
1. compare the performances and costs of currently available validated molecular DR tests and select one or more for use in the molecular DR testing service;
2. assess and overcome potential obstacles and barriers to a regional approach to diagnostic testing such as local regulations regarding out-of-state testing, reporting requirement, and need for memoranda of agreement;
3. develop a strategy to coordinate or integrate services provided by the molecular DR testing laboratories and the TB genotyping laboratories to avoid unnecessary duplication of efforts and shipment of isolates;
4. develop a roll-out strategy for the molecular DR testing service to include informing potential service users of the availability of the service, how to access the service, and the appropriate use and interpretation of molecular DR tests for TB;
5. define the scope of the molecular DR testing service more precisely by providing reliable estimates of the anticipated numbers of non-processed specimens, processed specimens, and cultures to be tested;
6. design a molecular DR testing service to meet the needs of local and state TB control programs; and
7. develop, compete, and award a request for contract to provide the services.
In addition to molecular DR testing, the resources of the molecular DR testing laboratory might be leveraged to provide other services for state and local TB programs and laboratories, such as NAA testing for detection, culturing, second-line drug susceptibility testing, or genotyping. The scope of any additional service must be clearly defined and adequately funded.
Cost estimate for 2500 samples per year (10 per day)
One laboratory needed for judicious use; a second laboratory may be needed as a backup. Four laboratories are likely to be needed for universal testing.
Estimates do not include the cost of processing specimens at the molecular DR testing laboratory.
CDC: Shipping of isolates to molecular DR testing laboratory ($27 per isolate) up to $67,500
Program: Shipping specimens to testing laboratory ($10 per specimen) up to $50,000
Molecular DR testing laboratory:
Consumables for molecular DR tests ($10 to $30 per sample tested) $25,000–75,000
Miscellaneous laboratory supplies ($10 per sample tested) $25,000
Personnel for (10–20 samples per day; 2500-5000 per year) $200,000
(1 FTE technician, 0.5 FTE data entry clerk, 0.5 FTE laboratory manager)
Overhead (amount depends on site) estimate 20% $50,000–60,000
Molecular DR testing laboratory subtotal: $300,000–360,000
Initial equipment (amount depends on method)
CDC Responsibilities
1. Develop a request for contract (RFC) that specifies the duties and responsibilities of the regional molecular DR testing laboratories and the proposal evaluation criteria. Advertise and compete the RFC. Review applications and select successful offerors. Provide funds.
2. Develop and fund a process (e.g., ‘bill-to-recipient’ FedEx account) for shipping M. tuberculosis cultures to the regional molecular DR testing laboratories.
3. Develop and fund a process for shipping specimens to the regional molecular DR testing laboratories for public health laboratories or programs that can not afford the cost of shipping specimens.
4. Provide guidance, templates, or models to assist TB Programs to develop criteria for selecting TB suspects or patients for molecular DR testing and develop and implement protocols for accessing the regional molecular DR testing services.
5. Work with molecular DR testing laboratories to develop standardized reporting.
6. Work with molecular DR testing laboratories to develop an external quality assurance, proficiency testing, or rechecking program for the molecular DR testing laboratories.
7. Develop a robust process for monitoring and evaluating the performance of the molecular DR testing laboratories. This should include post-market surveillance of the molecular DR testing to determine the performance, cost, and benefit of the molecular DR testing in a high-throughput setting.
8. Work with regional laboratories and programs to develop protocols to analyze discrepancies in the results of molecular and conventional DS tests. Serve as a referee laboratory to analyze discrepant results. Collect data on and investigate all discrepancies to better understand the performance of molecular and conventional DS testing.
9. Provide additional molecular and conventional DS testing for samples determined to be rifampin resistant.
10. Provide technical assistance and consultation.
Regional Molecular DR Testing Laboratory Responsibilities
1. Design and implement a molecular DR testing service to detect rifampin resistance and isoniazid resistance. The service should include
a. protocols and standard forms for submitting specimens for testing,
i. for universal testing, acceptable specimens should include non-processed respiratory specimens from TB suspects or patients who produced an AFB smear-positive or NAA positive specimen, processed AFB smear-positive respiratory specimens (sediments) from TB suspects or patients, and AFB-positive cultures from TB suspects or patients;
ii. specimens (respiratory or non-respiratory) or cultures identified by the TB program as high priority for molecular DR testing are acceptable.
b. protocols for accessioning samples and entering data in a laboratory information management system;
c. an algorithm and protocols for molecular DR testing of specimens and cultures;
i. include appropriate positive and negative controls
ii. ensure that testing includes the assessment of PCR inhibitors
iii. if inhibitors detected or suspected, appropriate follow-up testing
iv. if results are indeterminate, appropriate follow-up testing
v. if an open NAA system is used, protocols to prevent end-product contamination
d. procedures for result review and reporting - system must maintain patient confidentiality;
e. a quality management system and an external quality assurance program; and
f. procedures for archiving samples.
2. Preferably provide six-day-a-week testing.
3. Select, establish and validate rapid molecular test(s) to detect rifampin resistance and isoniazid resistance in accord with pertinent CLIA and FDA rules and regulations.
4. Conduct testing and report results of molecular DR testing within 2 business days of specimen receipt for at least 90% of samples received.
5. Work with programs to develop and implement protocols for reporting molecular DR test results. At the discretion of the program, this may include reporting to the TB Program, TB public health laboratory, local public health officials, sample submitter, etc. Results should be reported electronically to individuals designated by TB programs. The detection of resistance should be reported by telephone to individuals designated by the TB program.
6. Work with each TB program to develop and implement a reflex testing protocol to expedite testing of susceptibilities to first-line and second-line drugs for all rifampin-resistant samples.
a. The testing may be done in the same facility as the molecular DR testing, in another laboratory proficient in drug-susceptibility testing, or the original referring laboratory.
b. In addition, all isolates found to be rifampin resistant should be referred to CDC for additional molecular and conventional DS testing.
7. Work with CDC and TB Programs to develop and implement a protocol for analyzing discrepant results.
8. Participate in an external quality assessment or proficiency testing program.
Tuberculosis Control Program Responsibilities
1. Develop and implement a protocol that enables health care providers to access the molecular DR testing services in their jurisdiction. Include criteria for selecting TB suspects or patients for testing. For universal testing, criteria should allow testing of one AFB smear-positive or NAA-positive respiratory specimen or M. tuberculosis culture from each TB patient or TB suspect. Criteria for high priority or judicious testing might include:
a. Persons at risk of having MDR-TB such as persons exposed to an MDR-TB case, from a population with a high rate of MDR TB, failing or failed therapy with first-line anti-TB drugs, persons who have been previously treated for TB and relapse cases.
b. Specimens or isolates for which the result would alter case management or TB Control decisions, outbreak or contact investigations, preventive therapy, infection control, etc.
c. High profile situations needing test results in a short time frame (e.g. outbreaks in schools or congregate settings; placing persons on a ‘Do Not Board’ list).
d. Specimens or isolates that can not be tested easily with conventional methods such as non-viable specimens and mixed or contaminated cultures.
2. Ensure that health care providers are aware of the molecular DR testing service and protocol for accessing the service.
3. Coordinate programmatic and laboratory activities:
a. optimize communication between program, laboratory, and clinicians,
b. ensure that the public health laboratory is engaged early in the testing process for samples being submitted by other laboratories to facilitate referral and tracking of samples and expediting submission of samples to the public health laboratory, and
c. ensure that the public health laboratory is informed of all positive mol DS test results regardless of submitter to facilitate and expedite follow-up drug-susceptibility testing.
4. Develop protocols for approving requests for molecular DR testing.
5. Develop protocols for shipping specimens to the molecular DR testing laboratory. Work with CDC or specimen submitters to arrange for payment of costs, or pay for shipping costs if necessary.
6. Develop a protocol for shipping cultures to the molecular DR testing laboratory. Develop protocols for shipping specimens to the mol-DS testing laboratory. Work with CDC or specimen submitters to arrange for payment of costs, or pay for shipping costs if necessary
7. Designate individuals to be notified of test results or subsets of test results (e.g., negative test results, detection of M. tuberculosis DNA, detection of drug resistance, etc.). As appropriate, develop protocols to act upon test results.
8. Work with the molecular DR testing laboratory and CDC to develop and implement a reflex testing protocol that ensures prompt culture-based testing of susceptibilities to first-line and second-line drugs for all rifampin-resistant samples. Work with CDC and testing laboratories to arrange payment of the costs of the reflex testing. Cost to be paid by the TB program. For example, all rifampin-resistant isolates could be tested for susceptibility to first-line and second-line drugs at 1) the molecular DR testing facility, 2) the program’s public health laboratory, or 3) at another laboratory proficient in drug-susceptibility testing.
9. Develop and implement a protocol for detecting and analyzing discrepancies in the results of molecular and conventional DS tests. Work with CDC and the molecular DR testing laboratory to evaluate discrepancies.
10. Provide technical assistance and consultation on the appropriate use and interpretation of molecular DR tests to health care providers in their jurisdiction.
Local and State Public Health Laboratories
1. Work with local and state TB Control Programs to ensure that clinical laboratories in their jurisdictions are aware of the molecular DR testing service and protocol for accessing it.
2. Coordination of programmatic and laboratory activities.
3. Develop protocols with TB Control Programs for shipping specimens or isolates to the molecular DR testing laboratory.
4. Develop protocol with TB Control Programs to detecting and analyzing discrepancies in the results of molecular and conventional tests.
5. Work with TB Control Programs and the molecular DR testing laboratory to develop and implement a reflex testing protocol that ensures prompt culture-based testing of susceptibilities to first-line and second-line drugs.
6. Work with programs to develop and implement protocols for reporting molecular DR test results.
7. Provide technical assistance and consultation on the interpretation of molecular DR tests to laboratorians and health care providers in their jurisdiction.
Regional Training and Medical Consultation Center Responsibilities
1. Provide technical assistance and consultation on clinical and programmatic aspects of the appropriate use and interpretation of molecular tests for drug resistance.
2. Provide medical consultation as appropriate.
3. Develop and disseminate education material on molecular drug-resistance testing.
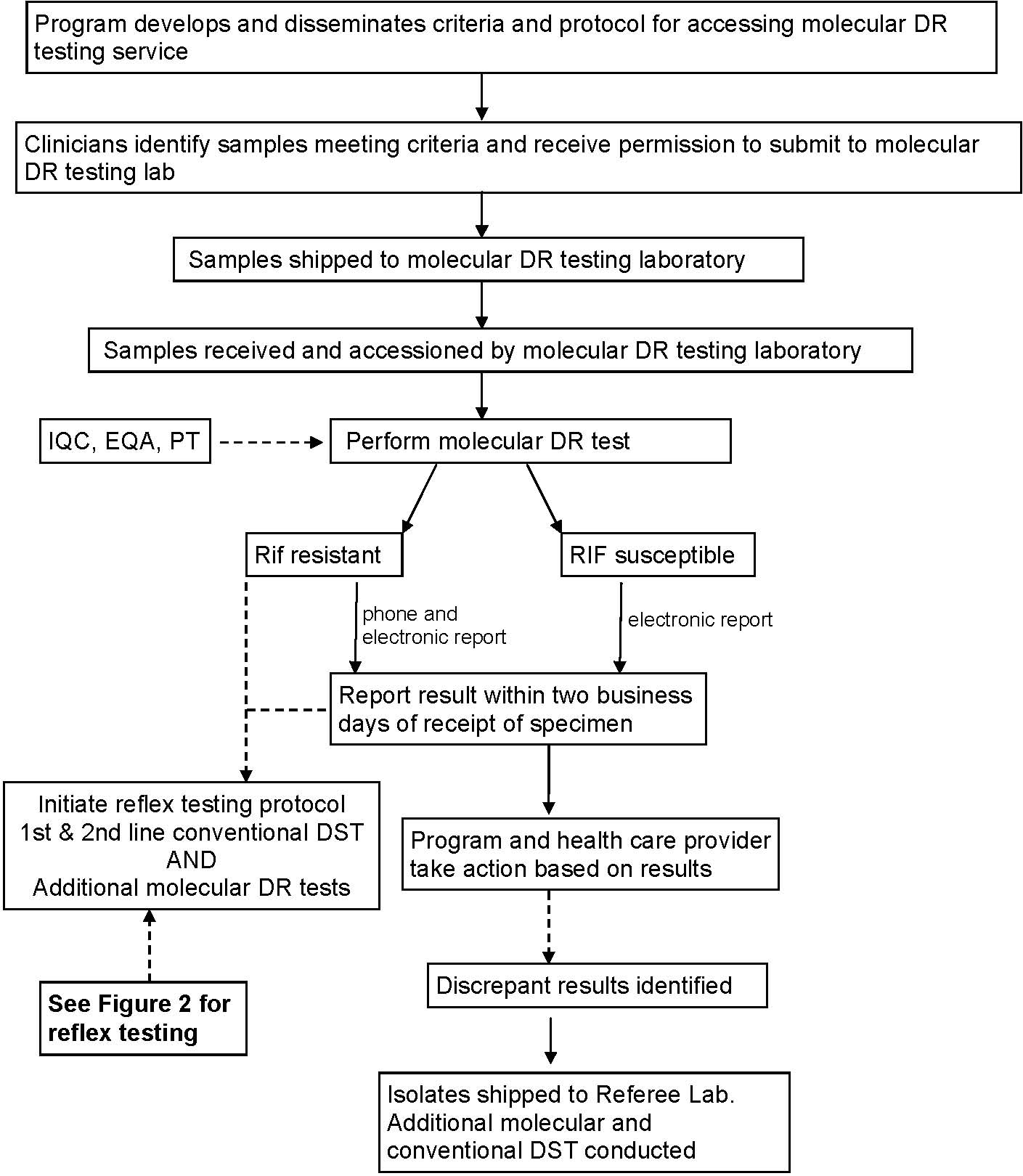

| File Type | application/vnd.openxmlformats-officedocument.wordprocessingml.document |
| File Title | Regional Laboratories for Molecular Drug Susceptibility Testing |
| Author | tms1 |
| File Modified | 0000-00-00 |
| File Created | 2021-01-31 |
© 2026 OMB.report | Privacy Policy