IC Justification
IC-PR DW Study.docx
Generic Clearance for Citizen Science and Crowdsourcing Projects (Renewal)
IC Justification
OMB: 2080-0083
Request for Approval under the “Generic Clearance for Citizen Science and Crowdsourcing Projects” (OMB Control Number: 2080-0083)

TITLE OF INFORMATION COLLECTION: Using Citizen Science to Improve Drinking Water Epidemiology Studies
PURPOSE: This project showcases the simplicity of an innovative saliva test and improves the way epidemiology studies are designed using citizen science. Families with school age children will report incidences of gastrointestinal disease to school nurses and/or science teachers to facilitate follow-up stool and saliva tests by EPA in impacted school districts. Drinking water, stool and saliva samples will be collected by researchers and analyzed by Interamerican University, EPA Office of Research and Development (ORD) and Region 2 scientists. The project will allow citizens to investigate the incidence and type of gastrointestinal illness in rural communities in Puerto Rico without municipal (PRASA) drinking water treatment plants. This will better characterize and inform public health concerns related to drinking water treatment processes.
NEED AND AUTHORITY FOR COLLECTION: InterAmerican University received an Institutional Review Board (IRB) for Protection of Human Subjects in Research on January 26, 2018. On April 5, 2018 this project received approval from EPA’s Human Subjects Research Review Office for use of Human Subjects Research according to the requirements of EPA Order 1000.17 Change A1 (Policy and Procedures on Protection of Human Research Subjects) and can confirm that this study complies with EPA Regulation 40 CFR 26 (Protection of Human Subjects).
USES OF RESULTING DATA: Results from all aspects of the epidemiology study will be compiled and interpreted for publication in peer-reviewed journal articles. The resulting data may provide a direct link between community health and drinking water treatment using citizen science in underserved communities in rural Puerto Rico.
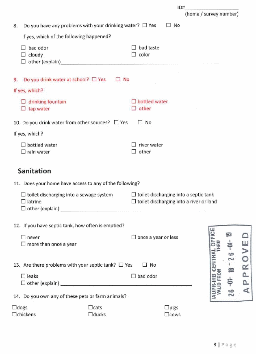
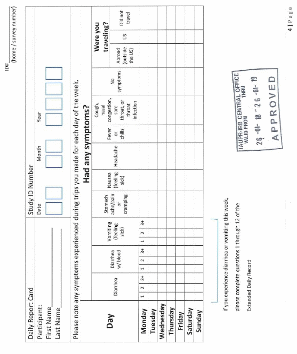
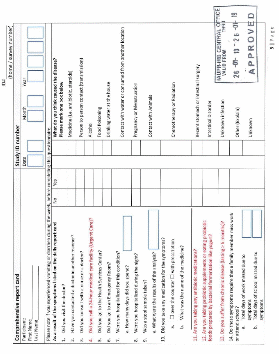
DATA COLLECTION METHODS: School nurses, science teachers, and/or researchers from EPA Region 2 and InterAmerican University will recruit and enroll local families with at least one child in the 4th to 6th grade. An adult family member or guardian will complete one baseline survey to gather limited demographic data and information about risks related to waterborne illness (i.e. water usage and sanitation). At approximately one-month intervals, for a total of 3 months, an adult family member or guardian will complete a health survey on symptoms experienced by the child participant. Completed surveys will be returned to school nurses and science teachers by the child participant or family member. Study staff from InterAmerican University will collect and compile surveys from schools. EPA and InterAmerican University will analyze stool, saliva and drinking water samples. Compiled survey information and analytical results will be interpreted to prepare a summary report that includes all aspects of the epidemiology study.
PARTICIPANT UNIVERSE:
Category of Respondent |
No. of Respondents |
Number of responses per respondent |
Participation Time per response |
Burden Hours |
Family members |
500 |
4 (1 baseline + 3 health surveys) |
15 minutes |
500 hours |
Totals |
500 |
|
|
500 hours |
AGENCY COST: The estimated cost to the Federal government is $70,000 for project design and implementation. $30,000 of sampling supplies will be purchased by EPA ($15,000/year). Four EPA employees are collaborating on this project. It is estimated that each spends about 20% of their time (0.20 FTEs each) on this citizen science project.
STATISTICAL ANALYSIS:
The objective of statistical analysis is to assess and compare associations between water quality and infections with specific potentially waterborne pathogens. Anticipated survey results will satisfy the survey objectives.
Two approaches to statistical analysis of assay data will be used. For acute infections which have a relatively short incubation period and produce short-term immunity, such as noroviruses, Campylobacter spp. and Cryptosporidium spp., immunoconversion will be used as an indicator of incident infection as described previously (Griffin et al., 2015). Immunoconversion will be defined as at least four-fold increase in salivary antibody response between consecutive samples. Additional criteria may be used to improve the specificity of the immunoconversion tests, such as age-specific cut-off values derived from regressing antibody data on age using penalized splines as described previously (Egorov et al., 2010), and at least three-fold increase in third sample (S3, collected 2 months after baseline) compared to baseline (S1) sample (Wade et al., manuscript in review). For chronic infections, H. pylori and T. gondii, analysis will focus on identifying chronically infected individuals. DNA-based molecular methods will aso be conducted to determine the bacterial diversity and the presence of waterborne pathogens in the stool samples.
Analysis of associations between water quality and acute infections will be conducted using logistic regression models. Analysis will be repeated for asymptomatic infections (immunoconversion, no symptoms) and symptomatic infections (immunoconversion with symptoms).
For chronic infections, demographic, socioeconomic and behavioral risk factors for infections will be explored. In addition, potential impacts of chronic infections on antibody responses to incident acute infections will be explored.
DATA QUALITY ASSESSMENT PROCEDURES:
All samples from the same individual will be assayed at the same time to minimize assay variability. Samples from at least 20% of study participants will be assayed in duplicate. Controls (human samples positive to pathogens included in this study) as well as negative controls (blanks) will be assayed on each 96 well microplate. All analytical errors, such as insufficient number of Luminex beads (less than 50 beads of each type) acquired by the Luminex device, will be documented. All samples associated with errors as well as all other samples from the same individuals will be re-analyzed on a new plate. Plates with unusually high antibody responses to controls (GST or total IgG) will be identified using analysis of distributions of plate-specific responses at the end of the study. Plates with antibody responses to internal control antigens above the mean plus two standard deviations (outliers) will be re-analyzed.
DNA extractions will be conducted using established protocol and following manufacturer’s instructions. Sequencing of the 16S rRNA will be used following the procedures outlined in Caporaso et al (2011) and as modified by Kapoor et al (2016). qPCR assays will follow the steps described in Kapoor et al (2015). Standards (dsDNA) will be used for quantifying gene copies and no template controls will be used to determine the presence of cross-contamination.
ADMINISTRATION OF THE INSTRUMENT: (Check all that apply)
[X] Web-based or Social Media
[X] Telephone
[X] In-person
[X] Mail
[ ] Other, Explain
INSTRUMENT: Instrument script is attached below. Final online product will include mandatory OMB control number, expiration date, and burden statement.
CONTACT NAME: __Craig Patterson____ EMAIL: ____ [email protected]_____
REFERENCES:
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108:4516-4522
Egorov, A.I., T.L.M. Montouri, L. Ascolillo, H.D. Ward, D.A. Levy, R.D. Morris, E.N. Naumova, J.K.
Griffiths. 2010. Recent diarrhea is associated with elevated salivary IgG responses to Cryptosporidium in residents of an eastern Massachusetts community. Infect., 38(2): 117-23.
Griffin, S.M., R.R. Converse, J.S. Leon, T.J. Wade, X. Jiang, C.L. Moe, A.I. Egorov. 2015.
Application of salivary antibody immunoassays for the detection of incident infections with Norwalk virus in a group of volunteers. J. Immunol. Methods, 424: 53-63.
Kapoor, V., Elk M., Li X., Impellitteri C.A., Santo Domingo J.W. 2016. Effects of Cr (III) and Cr (VI) on nitrification inhibition as determined by SOUR, function-specific gene expression and 16S rRNA sequence analysis of wastewater nitrifying enrichments. Chemosphere 147:361-367.
Kapoor , V., Ryu H, Pitkanen T, Wendell D, and Santo Domingo JW. 2015. Distribution of human-specific Bacteroidales and fecal indicators in an urban watershed impacted by sewage pollution using RNA and DNA based quantitative PCR assays. Applied and Environmental Microbiology. 81:91-99.
Wade, T.J., S.A.J. Augustine, S.M. Griffin, E.A. Sams, K.H. Oshima, A.I. Egorov, A.P. Dufour.
2017. Asymptomatic norovirus infection associated with swimming at a tropical beach. PLOS ONE, submitted.



| File Type | application/vnd.openxmlformats-officedocument.wordprocessingml.document |
| File Modified | 0000-00-00 |
| File Created | 0000-00-00 |
© 2026 OMB.report | Privacy Policy